Membrane Permeability
Membrane permeability quantifies how easily a molecule can pass through a given lipid membrane. This matters a lot for therapeutics:
- If a drug is taken orally, then it must pass through the gut epithelial membrane to enter the bloodstream. Drugs with low membrane permeability will thus have low oral bioavailability.
- Once in the bloodstream, the drug must pass through cell membranes in order to reach intracellular targets.
- Drugs targeting the central nervous system must also pass through the blood–brain barrier, an additional membrane that protects the brain from potentially harmful xenobiotics.
In the words of veteran drug hunter Derek Lowe, "membrane permeability is one of the most basic properties that we look for in a drug." Accordingly, it's common for drug-discovery scientists to test permeability extensively in preclinical research. There are a lot of experimental assays that are used: cell lines like Caco-2 and MDCK are often used to measure permeability, as are artificial membranes like PAMPA (for more details, see this excellent analysis by Karel Berka and co-workers). However, all of these experimental assays require a sample of the synthesized compound and cost additional time & money, limiting the potential throughput.
Since permeability is so crucial, it's become common to predict drug permeability computationally before actually synthesizing or ordering new compounds. While computational models are never as accurate as actually running the experimental assay, they can be run quickly and cost-effectively on thousands or tens of thousands of compounds, allowing scientists to avoid molecules predicted to have bad permeability and spend their limited time and resources only on the most promising candidates.
Predicting Membrane Permeability in Rowan
Rowan offers two complementary membrane-prediction methods.
GNN-MTL: A Multi-Task Graph Neural Network from AstraZeneca
Rowan's first permeability-prediction method comes from Philip Ohlsson and co-workers at AstraZeneca, who recently published a paper in ACS Omega showing that multi-task learning could improve the performance of graph neural networks for permeability prediction. The model associated with this paper was trained on large amounts of internal data from AstraZeneca and is open source; we’ve added this model to Rowan.
Ohlsson and co-workers' GNN-MTL model directly predicts experimental Caco-2 apparent permeability values from SMILES strings. To run this model in Rowan, simply input your desired molecule as a SMILES string, select the "GNN-MTL" method, and submit the workflow. The prediction takes only seconds to run, and predictions are color-coded for easy qualitative interpretation.
Submitting a membrane-permeability workflow on cimetidine.
PyPermm: Physical Modeling of Passive Diffusion
Rowan also supports physics-based permeability modeling through PyPermm, a refactored version of PerMM from Andrei Lomize and co-workers. PyPermm computes free-energy profiles for membrane transit as well as predicting intrinsic permeability coefficients for five different membrane types (plasma, blood–brain barrier, Caco-2, black lipid membrane, and PAMPA).
To run PyPermm-based permeability predictions in Rowan, simply select the PyPermm method on the workflow submit screen. PyPermm also runs quickly and returns a color-coded breakdown of predicted permeability for different membranes, with an accompanying predicted free-energy profile.
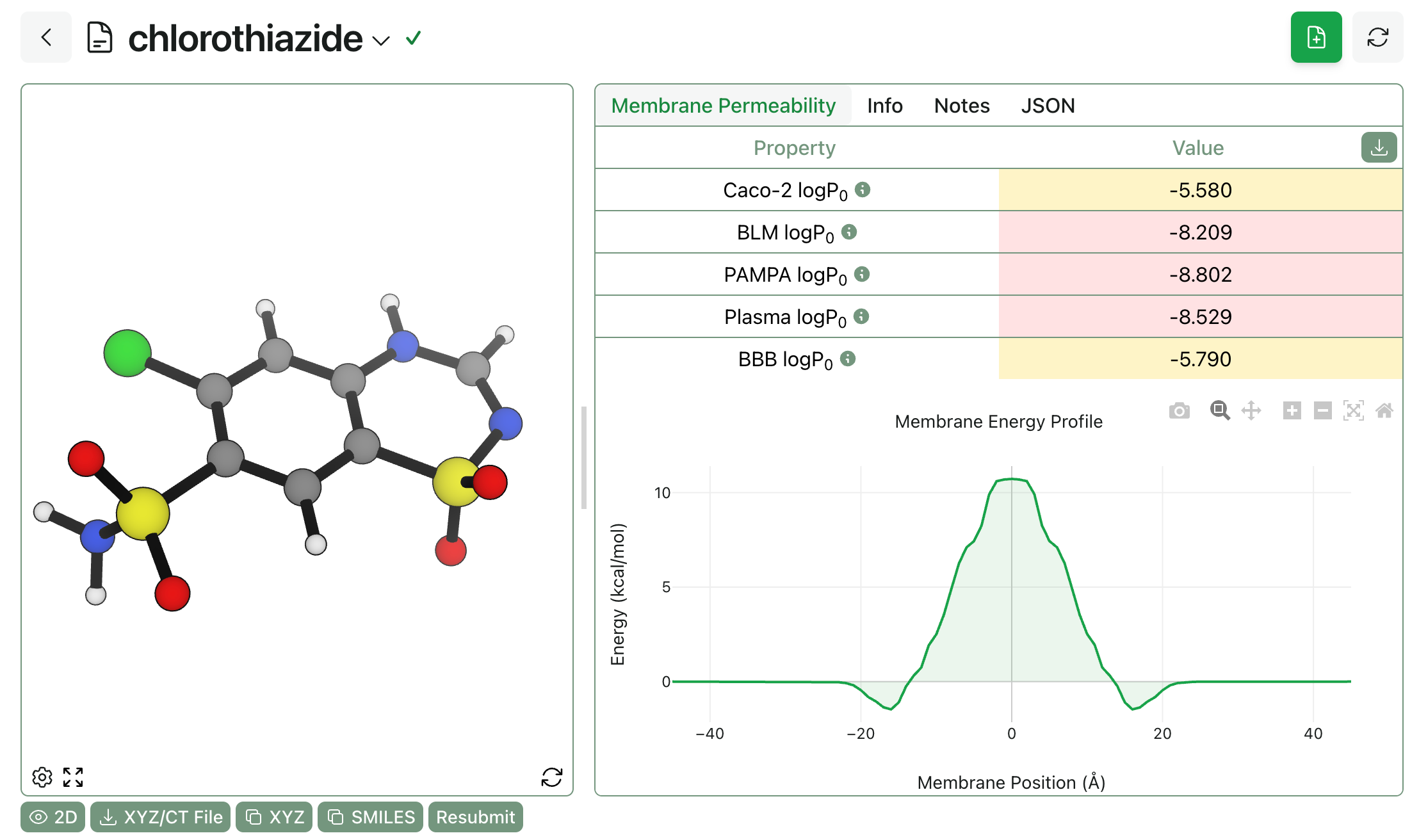
The output of a permeability workflow run on chlorothiazide.
Both GNN-MTL and PyPermm can also be run through Rowan's Python API:
from rdkit import Chem
import rowan
smiles = "CC1=C(N=CN1)CSCCNC(=NC)NC#N"
gnn_mtl_result = rowan.submit_membrane_permeability_workflow(
smiles,
name="GNN-MTL permeability",
)
pypermm_result = rowan.submit_membrane_permeability_workflow(
Chem.MolFromSmiles(smiles),
method="pypermm",
name="PyPermm permeability",
)
gnn_mtl_result.wait_for_result().fetch_latest(in_place=True)
pypermm_result.wait_for_result().fetch_latest(in_place=True)
print(f"Caco-2 Papp (GNN-MTL): {gnn_mtl_result.data['caco_2_P_app']}")
print(f"Caco-2 P0 (PyPermm): {pypermm_result.data['caco_2_logP']}")
