Predicting Ion-Mobility Mass Spectra
Ion-mobility mass spectrometry is a type of mass spectrometry (MS) that’s often used in drug discovery and metabolomics. In addition to measuring mass-to-charge ratio (m/z) like other MS techniques, ion-mobility MS also measures the collision cross section (CCS) of different ions. CCS values, which are measured in area (typically Å2), are exquisitely sensitive to molecular shape and conformation, making them useful for distinguishing structural or stereochemical isomers in complex mixtures.
Unfortunately, predicting the CCS value of unseen molecules remains a challenging computational task. Conventional physics-based methods require costly conformer optimization and scoring with DFT followed by trajectory simulations to determine the nitrogen cross section of a given molecular conformation, while ML-based methods to date have shown insufficient accuracy and generalizability for most real-world applications (at least according to this review).
Rowan's Approach to CCS Prediction
Rowan uses modern low-cost computational methods to generate physics-based CCS predictions in a fraction of the time that conventional workflows require. Rowan's workflow, documented in more detail in this technical blog post, uses CREST and GFN2-xTB to generate and optimize various conformers, g-xTB to score the output conformations, and a modified version of the open-source CoSIMS trajectory-simulation code to compute per-conformer CCS values. The final workflow can go from an input neutral analyte structure (specified as SMILES) to a final predicted CCS value in only a few minutes.
These CCS predictions are already being validated against real-world problems. In a early pilot study with the Gair Group at Michigan State University, MSU researchers found that Rowan's CCS predictions can be useful in quickly assigning isomeric mixtures that would otherwise require extensive isolation and characterization. Professor Joe Gair says:
Rowan has opened new research directions for our group. Comparing collisional cross sections calculated in Rowan versus those measured by ion-mobility mass spectrometry, we can assign structures to mixtures of diastereomers or regioisomers in an MS experiment that takes seconds.
Predicting CCS Through Rowan
Subscribing Rowan users and organizations can run CCS predictions through the web-based GUI. Simply navigate to the ion-mobility mass spectrometry workflow, input the desired molecule, and click "Submit"—Rowan automatically allocates compute resources and runs the entire workflow described above, with no additional intervention needed.
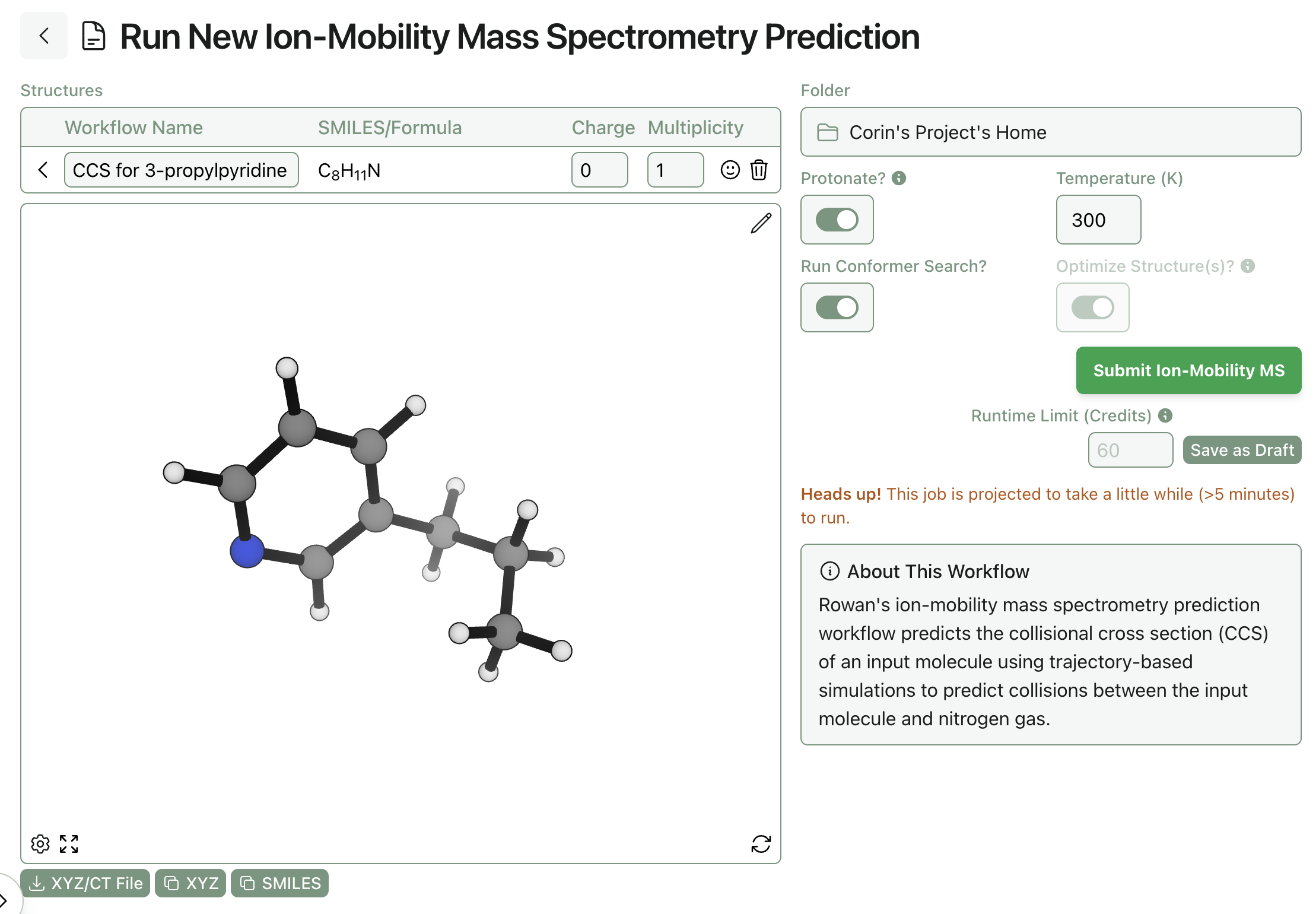
Submitting an ion-mobility MS prediction through Rowan.
The overall CCS prediction, alongside per-conformer predictions and weights, can be viewed on Rowan when the job is finished.
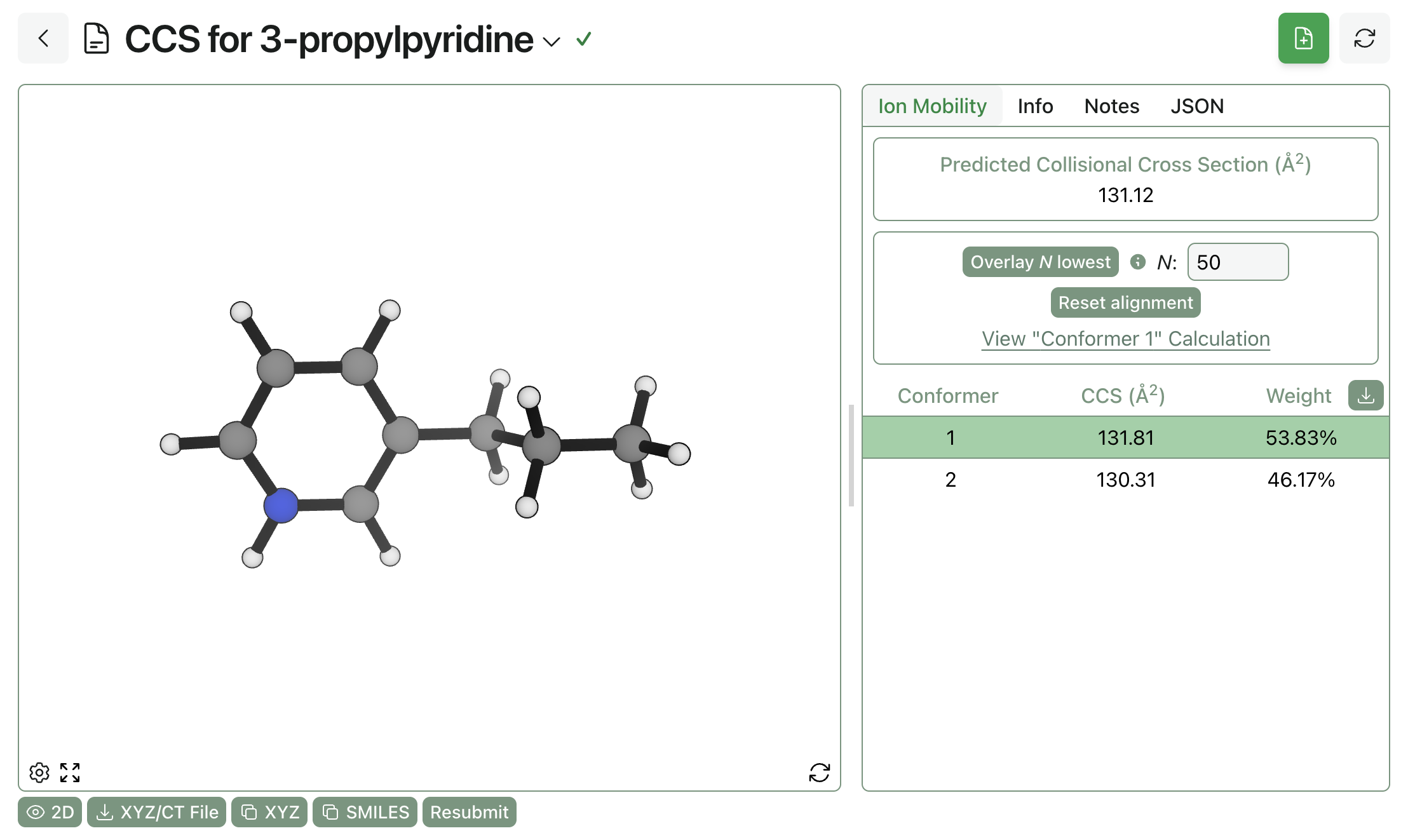
Viewing the result of an ion-mobility MS prediction through Rowan.
It's also easy to submit ion-mobility MS simulations through Rowan's Python API. Here's what an example prediction might look like:
import rowan
import stjames
workflow = rowan.submit_ion_mobility_workflow(
stjames.Molecule.from_smiles("c1c(COC)nccn1"),
protonate=True
)
workflow.wait_for_result()
workflow.fetch_latest(in_place=True)
avg_ccs = workflow.data["average_ccs"]
print(f"Predicted CCS: {avg_ccs:.2f} Å**2")
