Lyrebird: Molecular Conformer Ensemble Generation
by Eli Mann · Nov 5, 2025
This work was conducted by Vedant Nilabh, a summer intern from Northeastern University. Thanks Vedant!

Most molecules can exist in different 3D shapes, called conformers. Each conformer is a local minima on the potential-energy surface and has an associated energy, which determines its population at a given temperature. The observed macroscopic behavior of a molecule typically arises in part from all relevant conformations, making proper conformer search and ranking an important part of almost all chemical simulation problems.
Unfortunately, finding all the conformers of a given molecule is very difficult. There are a variety of commonly used methods, each with its strengths and limitations. At Rowan, we've generally relied on two methods to date: ETKDG, a stochastic distance-geometry-based approach incorporating experimental torsional heuristics, and CREST, an iterative metadynamics-based approach that also incorporates a genetic-structure-crossing algorithm to increase diversity. While we've had great success with both of these methods (like many other groups), both have their problems—ETKDG is somewhat inaccurate, particularly for large and flexible molecules, and can fail in particularly complex cases, while CREST is extremely slow and often struggles to explore enough space in a reasonable time. As such, we've been on the lookout for alternative conformer-generation methods.
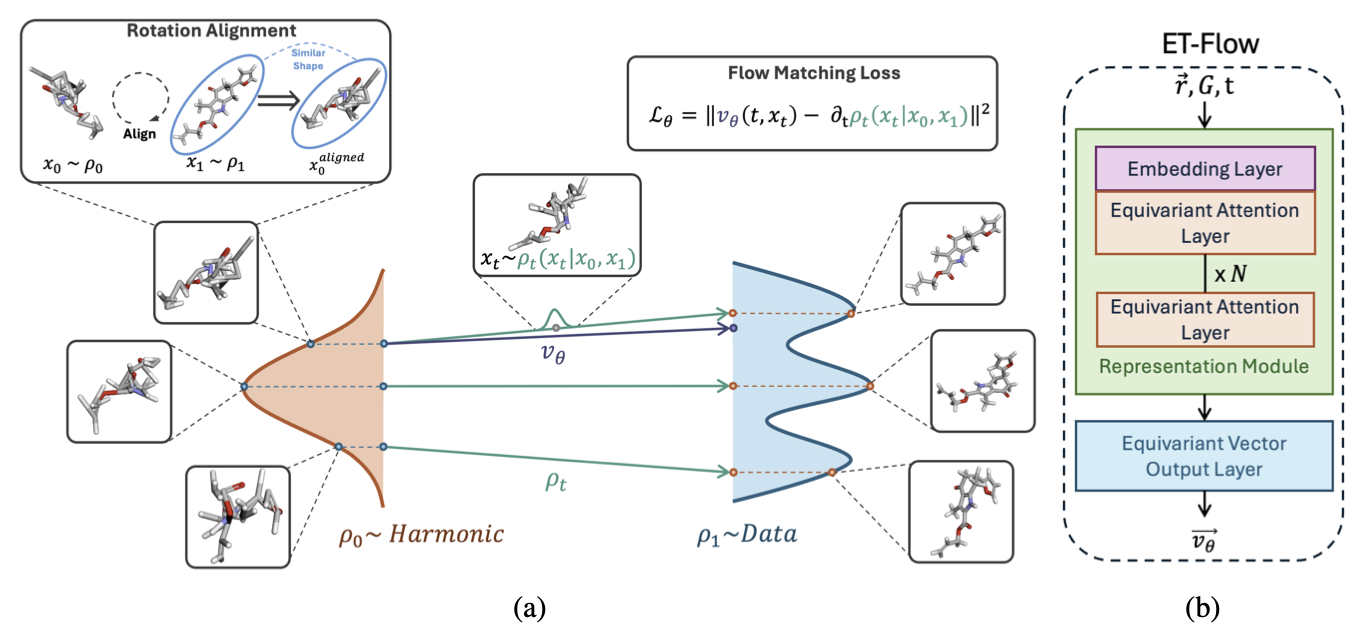
Lyrebird is our first foray into machine-learning-based conformer-generation algorithms. The Lyrebird architecture is based on the ET-Flow equivariant flow-matching architecture from Hassan et al. (preprint, GitHub). The model learns a conditional vector field that transports samples from a harmonic prior, conditioned on a covalent-bond graph, to the true distribution of 3D molecular conformers. The flow model then integrates a deterministic ODE to continuously transform these prior samples into realistic conformations. Because the network is SE(3)-equivariant, the learned vector field respects rotational and translational symmetries of molecules.
The original ET-Flow model was trained on a split of the GEOM-DRUGS subset of the GEOM dataset from Bombarelli et al., which contains over 317,000 ensembles of mid-sized drug-like organic molecules . Their studies show that the models perform well for molecules sampled within their training distributions, but poorly for molecules outside of their distribution. For Lyrebird, we increased the in-distribution samples by training on three datasets: GEOM-DRUGS; GEOM-QM9, a dataset with 133,258 small organic molecules limited to 9 heavy atoms; and CREMP, a dataset with 36,198 unique macrocyclic peptides. We hypothesized that increasing the diversity of the training dataset might lead to increased model generalizability, as well as improving the robustness of the model for routine chemical modeling tasks.
To test this hypothesis, we tested Lyrebird on Butina splits of GEOM-QM9, GEOM-DRUGS, and CREMP, as well as several challenging external sets: MPCONF196GEN, a small dataset containing conformers ensembles of the structures from MPCONF196, and GEOM-XL, a set of flexible organic compounds with up to 91 heavy atoms.
We evaluated our models against a variety of ML methods, as well as ETKDGv3, with metrics that evaluate both the diversity and geometric accuracy of a generated conformer ensemble. (We didn't benchmark against CREST because CREST was used to generate the training-data ensembles.) The metrics used for comparing conformer ensembles are a bit complex, because comparing two ensembles is a bit tricky, and merit specific explanation:
- Recall coverage reports the percentage of reference conformers that are successfully reproduced within a specified RMSD threshold δ.
- Recall AMR (Average Minimum RMSD) measures the average closest-match distance between each reference conformer and conformers in the generated ensemble; lower values indicate greater accuracy.
- Precision coverage measures the percentage of generated conformers that correspond to at least one reference structure, indicating how many generated samples are useful rather than redundant or incorrect.
- Precision AMR (Average Minimum RMSD) reports the closest-match RMSD from each generated conformer to the reference set, where lower values indicate that the generated conformers are not only diverse but also structurally faithful.
| Method | Recall Coverage ↑ (Mean) | Recall Coverage ↑ (Median) | Recall AMR ↓ (Mean) | Recall AMR ↓ (Median) | Precision Coverage ↑ (Mean) | Precision Coverage ↑ (Median) | Precision AMR ↓ (Mean) | Precision AMR ↓ (Median) |
|---|---|---|---|---|---|---|---|---|
| Torsional Diffusion | 86.91 | 100.00 | 0.20 | 0.16 | 82.64 | 100.00 | 0.24 | 0.22 |
| ET-Flow | 87.02 | 100.00 | 0.21 | 0.14 | 71.75 | 87.50 | 0.33 | 0.28 |
| RDKit ETKDG | 87.99 | 100.00 | 0.23 | 0.18 | 90.82 | 100.00 | 0.22 | 0.18 |
| Lyrebird | 92.99 | 100.00 | 0.10 | 0.03 | 86.99 | 100.00 | 0.16 | 0.05 |
Table 1: GEOM-QM9 test set results (threshold δ = 0.5 Å). Coverage in %, AMR in Å. Best results in bold.
| Method | Recall AMR ↓ (Mean) | Recall AMR ↓ (Median) | Precision AMR ↓ (Mean) | Precision AMR ↓ (Median) |
|---|---|---|---|---|
| RDKit ETKDG | 4.69 | 4.68 | 4.73 | 4.71 |
| ET-Flow | 4.13 | 4.07 | >6 | >6 |
| Lyrebird | 2.34 | 2.33 | 2.82 | 2.81 |
Table 2: CREMP test set results. Lower AMR is better (↓). Best results in bold. Coverage not reported because all methods have very low ensemble coverage.
| Method | Recall AMR ↓ (Mean) | Recall AMR ↓ (Median) | Precision AMR ↓ (Mean) | Precision AMR ↓ (Median) |
|---|---|---|---|---|
| RDKit ETKDG | 2.92 | 2.62 | 3.35 | 3.15 |
| Torsional Diffusion* | 2.05 | 1.86 | 2.94 | 2.78 |
| ET-Flow | 2.31 | 1.93 | 3.31 | 2.84 |
| Lyrebird | 2.42 | 2.07 | 3.27 | 2.87 |
Table 3: GEOM-XL test set results. Lower AMR is better (↓). Best results in bold. Coverage not reported because all methods have very low ensemble coverage.
*Torsional Diffusion generated only 77/102 ensembles.
| Method | Recall AMR ↓ (Mean) | Recall AMR ↓ (Median) | Precision AMR ↓ (Mean) | Precision AMR ↓ (Median) |
|---|---|---|---|---|
| RDKit ETKDG | 3.79 | 3.71 | 4.01 | 3.91 |
| Torsional Diffusion* | 2.71 | 2.58 | 3.13 | 2.95 |
| ET-Flow | 2.60 | 3.33 | 2.83 | 3.59 |
| Lyrebird | 2.54 | 2.96 | 2.80 | 3.56 |
Table 4: MPCONF196GEN test set results. Lower AMR is better (↓). Best results in bold. Coverage not reported because all methods have very low ensemble coverage. *Torsional Diffusion generated only 12/13 ensembles.
We found that Lyrebird outperforms ETKDG, in terms of both precision and recall, on every precision/recall metric we studied. Versus other ML methods like Torsional Diffusion and ET-Flow, the results are a bit more mixed—Lyrebird performs better when there's more relevant training data (e.g. Tables 1 and 2), but doesn't in general seem to generalize significantly better for "difficult" benchmark sets like GEOM-XL (Table 3) or MPCONF196GEN (Table 4). In general, all methods seem quite poor on these sets (an RMSD of 2.5 Å hardly inspires confidence).
We're excited to list the Lyrebird model on Rowan today for all users. While it's not a massive improvement over the previous ET-Flow method in areas similar to the core GEOM-DRUGS dataset, we anticipate that the increased diversity of the training data will make Lyrebird more robust and generalizable across the variety of scientific areas that our users study. As people use this model more, we look forward to seeing how well it performs on real-life use cases, particularly in comparison to existing methods like ETKDG and CREST. We note that Lyrebird is a newly released model, and that results should be carefully checked for production use cases before being relied upon—we don't expect that Lyrebird will be as reliable as ETKDG or CREST yet.
In parallel with this launch, we're releasing the Lyrebird weights on GitHub under an MIT license, making it easy for users to run Lyrebird locally or as a part of different workflows. We're also releasing our new MPCONF196GEN benchmark set under an MIT license for other groups to use when benchmarking conformer-generation methods.