Aqueous Solubility Prediction with Kingfisher and ESOL
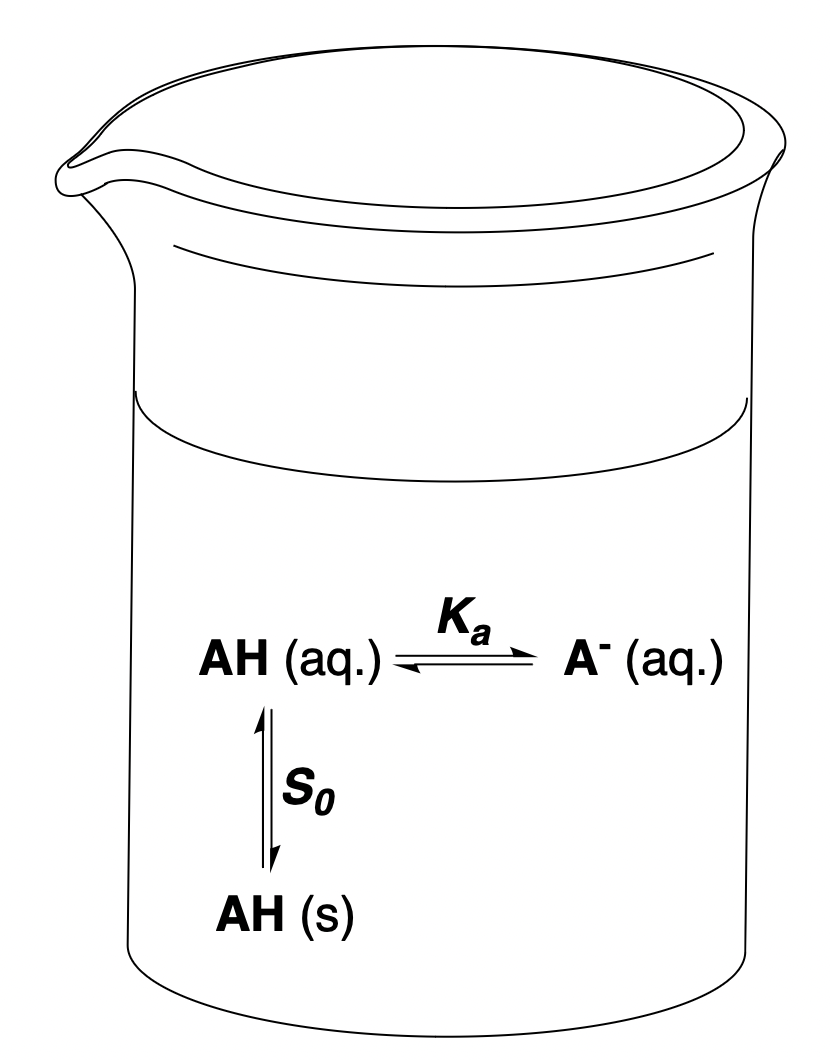
Aqueous solubility diagram of AH. Figure from our aqueous-solubility paper.
The aqueous solubility of a potential drug is an important factor in determining how well it will be absorbed into the bloodstream and reach its target. Thus, the accurate prediction of aqueous solubility for unseen organic molecules is a crucial tool in early-stage drug design.
Aqueous-solubility prediction is a notoriously difficult property to predict. Prior approaches have poor performance for unseen molecules and this remains an open problem in the field of cheminformatics. We studied aqueous solubility prediction methods from traditional multiple–linear regression approaches, to the cutting-edge pretrained neural network potential-based methods. We trained each of these models on the large, high quality Falcón-Cano et. al. "reliable" aqueous solubility dataset.
We also offer a method for predicting pH-dependent aqueous solubility, using Kingfisher and Starling, our ML-powered macroscopic pKa prediction model—a task which has previously been impossible due to the lack of large, publicly available, pH-dependent aqueous solubility datasets.
ESOL and Kingfisher Models
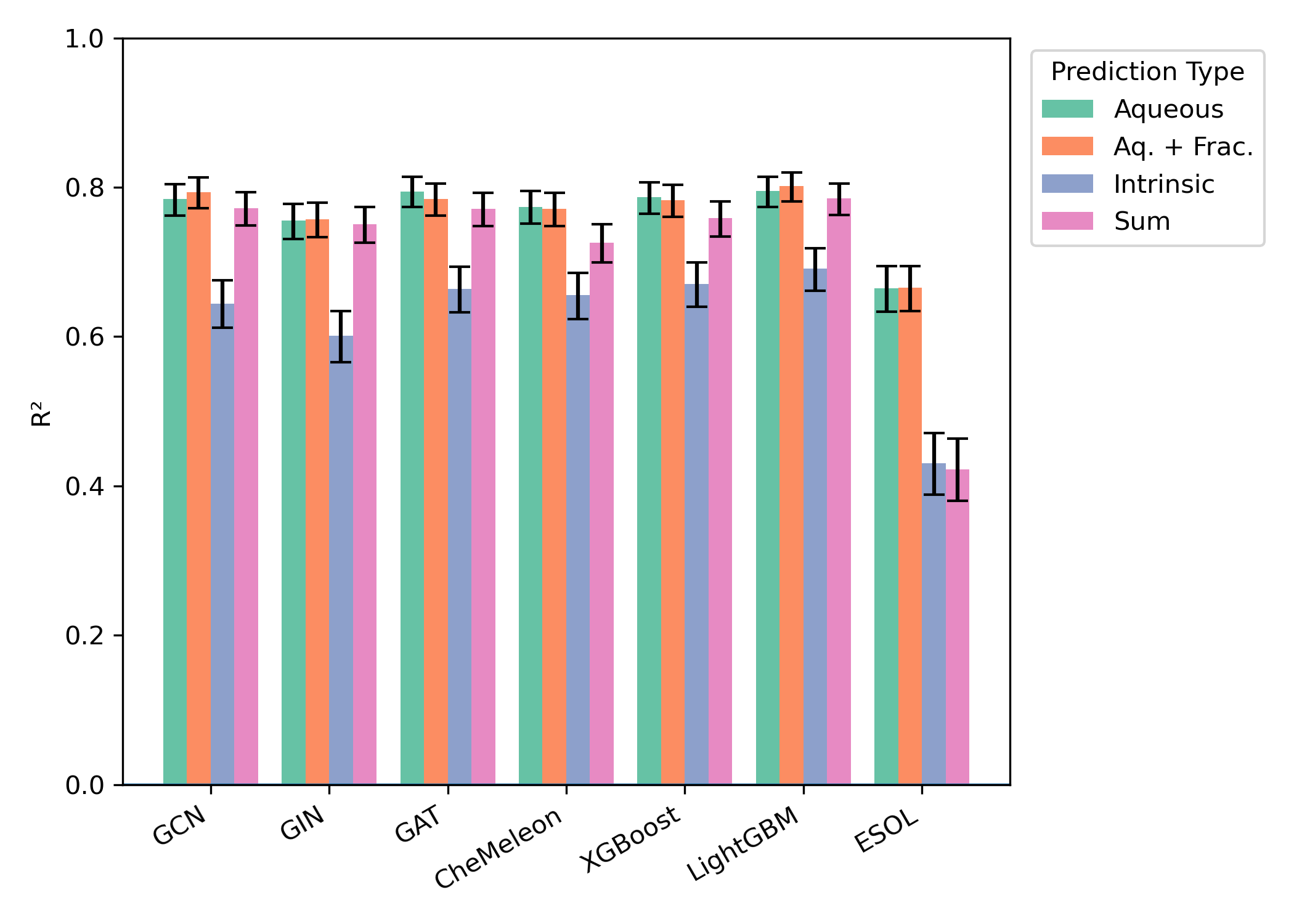
Model performance on 1,255 molecule Butina-split test set. Figure from our aqueous-solubility paper.
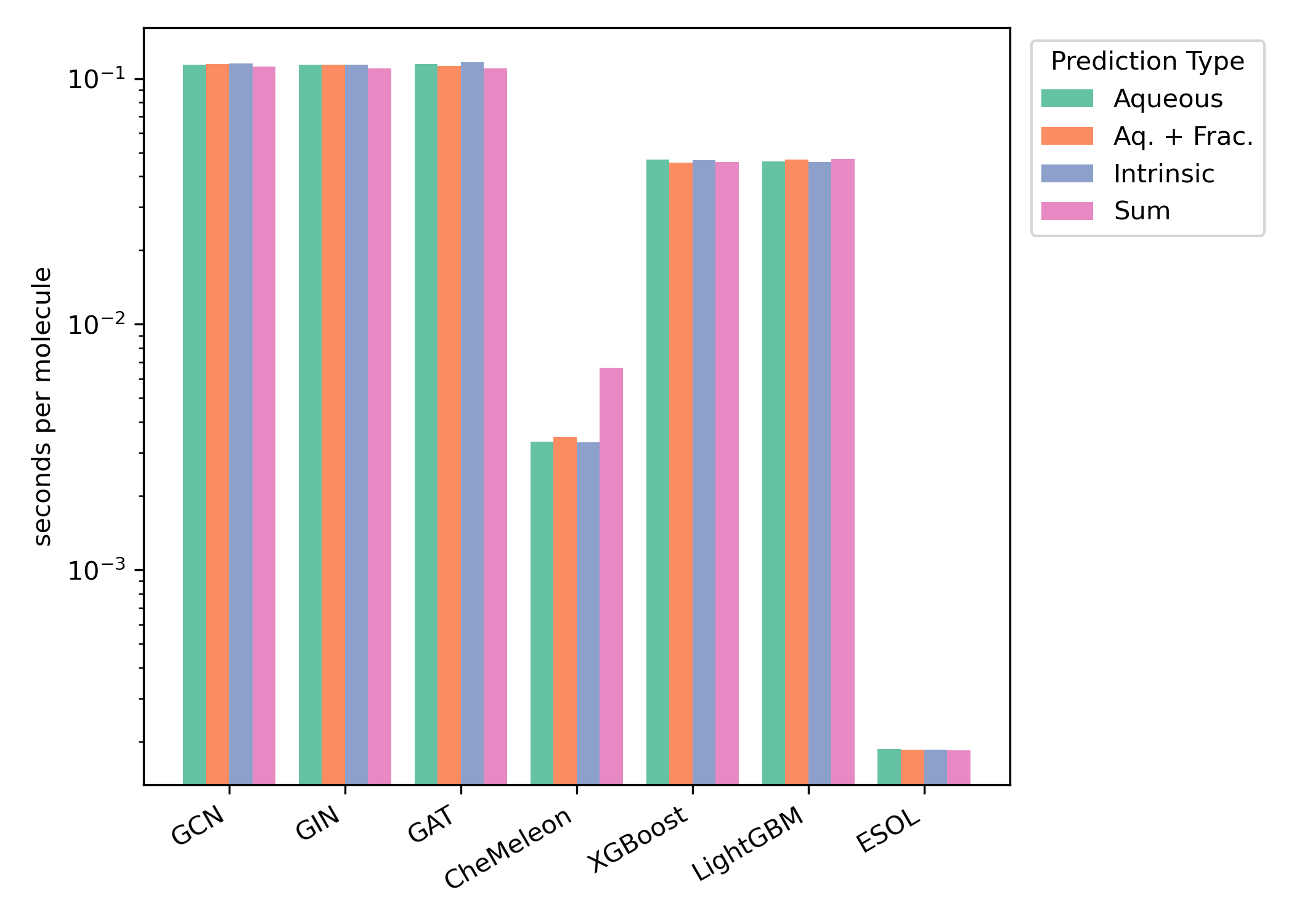
Average CPU inference time per-molecule Butina-split test set. Figure from our aqueous-solubility paper.
Based on our testing, we offer two models for aqueous solubility prediction on Rowan: a reparameterized ESOL and "Kingfisher."
ESOL is a multiple–linear regression model for aqueous solubility prediction developed by John S. Delaney at Syngenta. We reparameterized this model using an RDKit-based implementation from Pat Walters. This is a fast and trustworthy method which has been widely used for aqueous solubility prediction.
Kingfisher is a topological-molecular-connectivity-graph-based message-passing neural network. This model was built using the pretrained CheMeleon model from Jackson Burns and co-workers at MIT and fine-tuned on our chosen solubility dataset.
How pH-Dependent Solubility Prediction Works
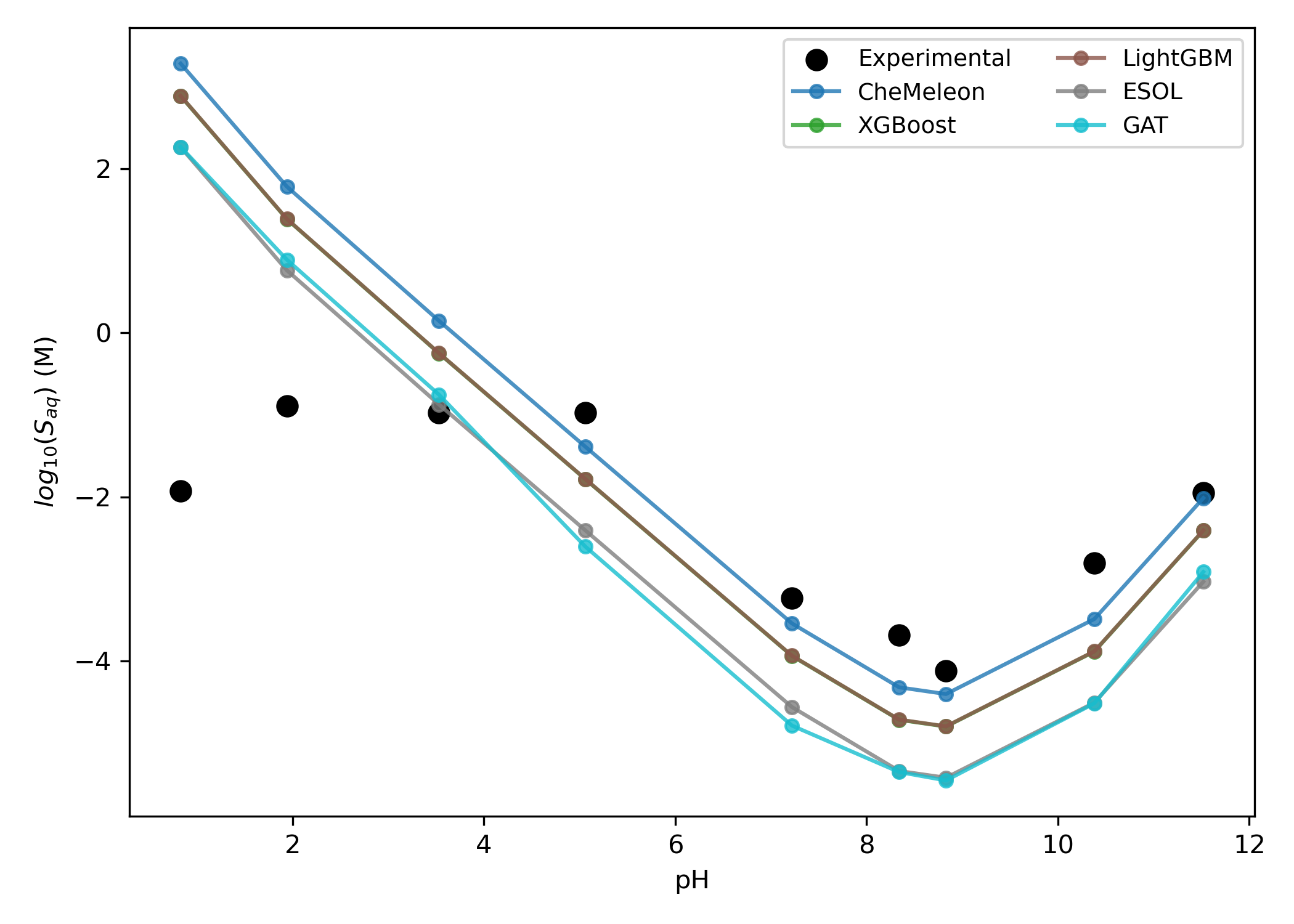
A comparison of strategies for pH-dependent aqueous solubility prediction. Figure from our aqueous-solubility paper.
Rowan predicts pH-dependent solubility by running a macroscopic pKa calculation, predicting the aqueous solubility at neutral pH with Kingfisher, and scaling by the fraction of neutral microstates at each pH. This generates pH-dependent solubility relationships with good accuracy, although non-ideal behavior like aggregation is not modeled through this framework.
How to Run Aqueous Solubility Calculations Through Rowan
Aqueous solubility can be predicted through Rowan's solubility workflow. Choose which solubility-prediction method you would like to use, choose the appropriate temperature and solvent for aqueous solubility, and submit.
An example of submitting an aqueous solubility prediction for ibuprofen.
pH-dependent aqueous solubility prediction can be run though our macroscopic pKa workflow. Ensure that "Predict pH-Dependent Aqueous Solubility?" is enabled before submitting the workflow. To view results, navigate the the "Aqueous Solubility" tab after the calculation finishes.
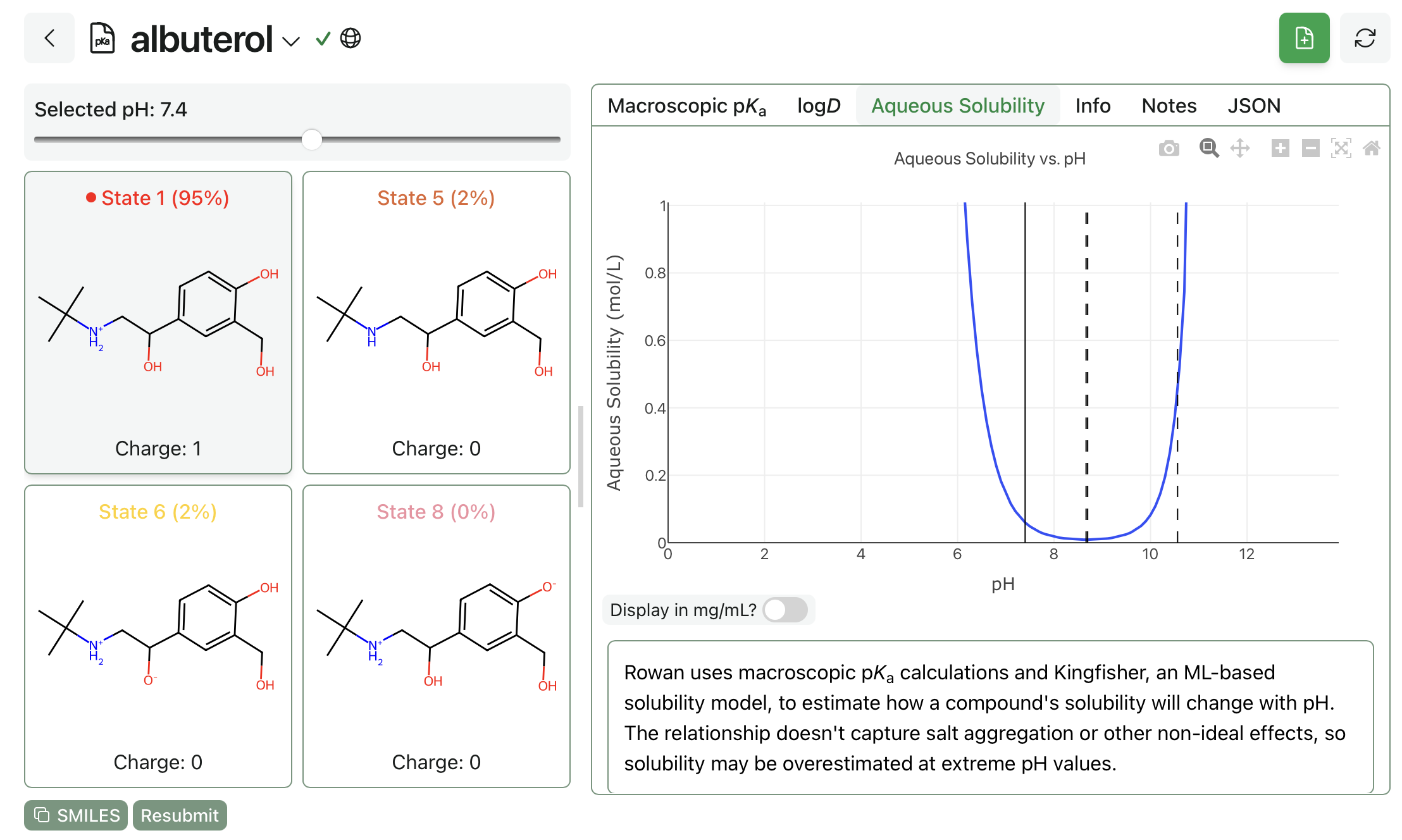
pH-dependent aqueous solubility prediction for albuterol.
