Double-Ended Transition-State (TS) Search
Double-ended transition-state (TS) search methods generate guess TS structures based on input reactant and product structures.
Running Double-Ended TS Searches
Double-ended TS searches start from reactants and products to generate a guess for the transition state connecting these structures. Currently, the Rowan platform supports the freezing-string method (FSM) for double-ended TS searches. FSM starts from reactant and product geometries and forms "frozen" strings of geometries that expand towards the opposite string end while attempting to stay within the reaction channel. Once the ends are connected, the peak along the path is chosen as a guess for the TS and optimized (learn more about double ended string methods in our Guessing Transition States blog).
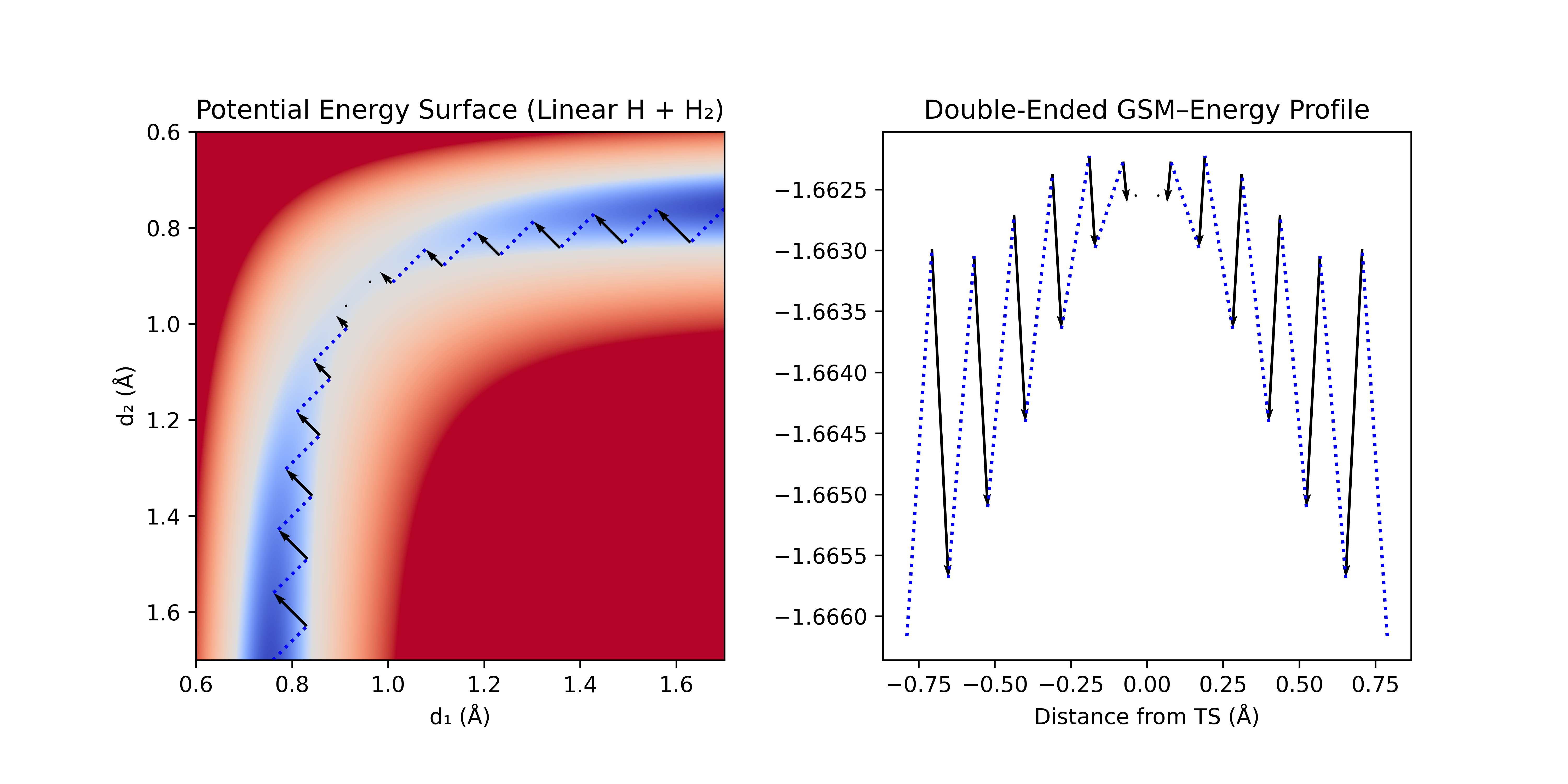
Figure 1: Double-ended FSM with linear synchronous transit interpolation. A step (dotted blue line) is taken along the linear interpolation towards the other end, and then optimized normal to the step (black arrow). This process is repeated until the ends meet.
Rowan's double-ended TS search workflow takes input reactant and product geometries and runs the freezing-string method to generate a guess for the transition state. The workflow can optionally optimize the reactant and product structures before running the double-ended TS search, as well as optimize the guessed transition state after the search is complete. The results are streamed to Rowan's web interface in real time, enabling easy monitoring, analysis, and resubmission from double-ended TS searches as they run. Rowan's web interface also allows for visualization of how coordinates change across the course of the double-ended TS search: simply select a geometric coordinate in the molecule viewer and Rowan will automatically plot how this variable evolves on the double-ended TS search graph.
Rowan's double-ended TS search workflow can be run with any method that supports gradients, including DFT, semiempirical, and NNPs (see below).
Supported Electronic Structure Methods and ML Potentials
API
Double-ended TS searches can also be run via the API, enabling high-throughput and simple integration into existing workflows.
import rowan
from stjames import Settings, Molecule, Method
from stjames.optimization.freezing_string_method import FSMSettings
from stjames.workflows.double_ended_ts_search import DoubleEndedTSSearchWorkflow
HCN = Molecule.from_xyz("""\
H 0 0 -1.1
C 0 0 0
N 0 0 1.2""")
CNH = Molecule.from_xyz("""\
H 0 0 2.3
C 0 0 0
N 0 0 1.2""")
workflow = DoubleEndedTSSearchWorkflow(
reactant=HCN,
product=CNH,
calculation_settings=Settings(method=Method.AIMNET2_WB97MD3),
search_settings=FSMSettings(),
optimize_inputs=True,
optimize_ts=True,
)
result = rowan.submit_workflow(
initial_molecule=HCN,
workflow_data=workflow.model_dump(),
name="HCN Isomerization",
workflow_type="double_ended_ts_search",
)
print(f"View workflow privately at: https://labs.rowansci.com/double-ended-ts-search/{result.uuid}")
result.wait_for_result().fetch_latest(in_place=True)
print(result.data["forward_string_distances"])
print(result.data["backward_string_distances"])
print("Forward calculations")
for uuid in result.data["forward_calculation_uuids"]:
print(f"- https://labs.rowansci.com/sub_calculation/{uuid}")
print("Backward calculations")
for uuid in result.data["backward_calculation_uuids"]:
print(f"- https://labs.rowansci.com/sub_calculation/{uuid}")
print(result)
