Intrinsic Reaction Coordinate (IRC)
Intrinsic reaction coordinate (IRC) calculations show the lowest-energy path connecting the reactants and products through a given transition state (TS). IRCs provide insight into reaction mechanisms and confirm the validity of a proposed TS (as it is not uncommon for a proposed transition state to lead to a different reaction than the one being targeted [such as an internal rearrangement or even a methyl rotation]).
Running IRCs
IRC calculations start from an optimized transition state (which contains a single imaginary frequency corresponding to the proposed reaction). The IRC takes a series of mass-weighted steps along the paths of steepest descent away from the TS in both a "forward" and "backward" direction until minima are found (or the maximum number of steps is exceeded).
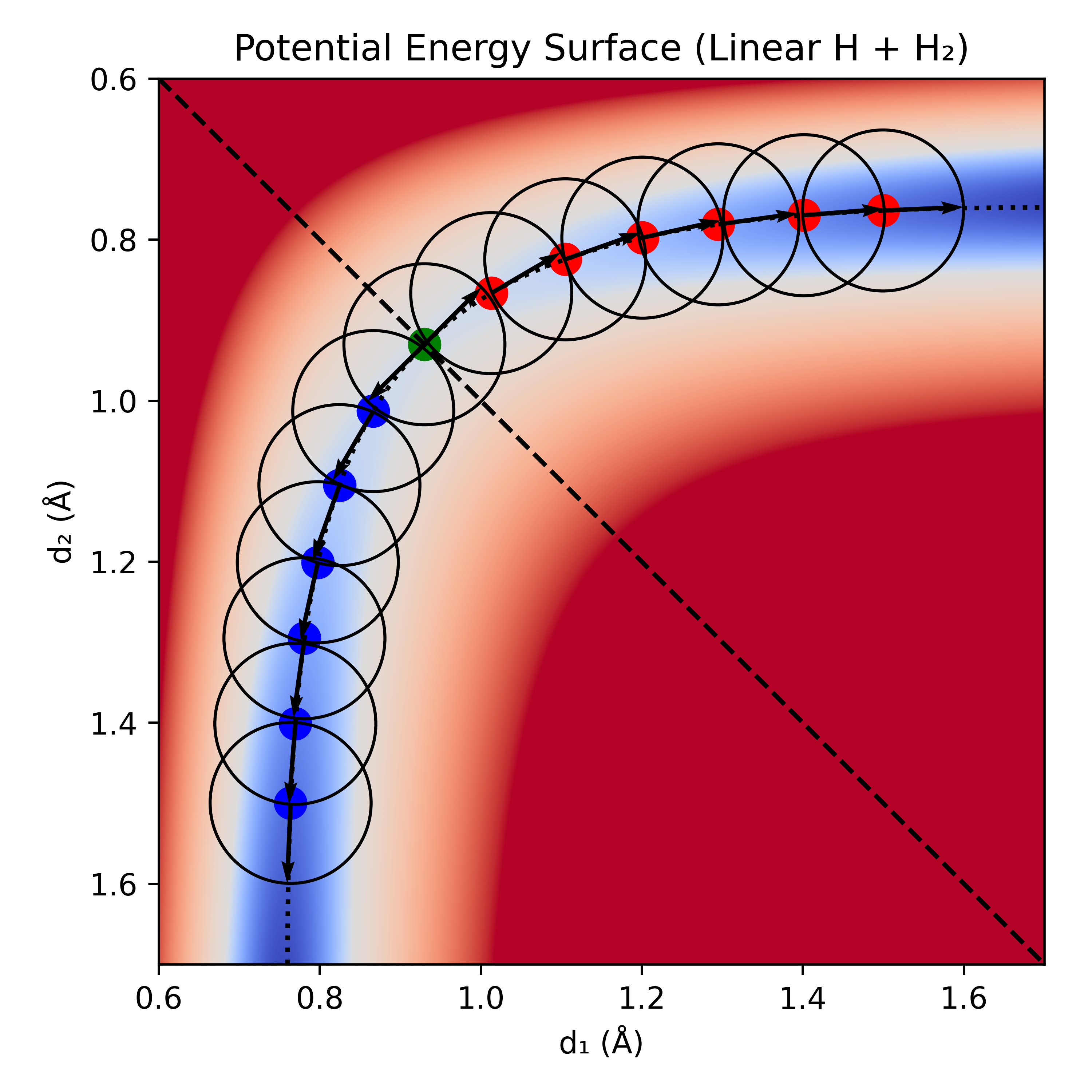
Sample IRC for the H + H2 reaction with six 0.1 Å·amu–1/2 steps in each direction.
Rowan's IRC workflow takes an input transition-state structure and runs the IRC in both the forward and backward directions. The results are streamed to Rowan's web interface in real time, making it easy to monitor, analyze, and resubmit from IRCs as they run. Rowan's web interface also allows for visualization of how coordinates change across the course of the IRC: simply select a geometric coordinate in the molecule viewer and Rowan will automatically plot how this variable changes on the IRC graph. All IRC data can also be downloaded as a .csv file.
Rowan's IRC workflow can be run with any method that supports gradients, including DFT, semiempirical, and NNPs (see below).
Supported Electronic Structure Methods and ML Potentials
API
IRCs can also be run via the free-to-use Python-native API, enabling high-throughput and simple integration into existing workflows.
from stjames import Molecule
import rowan
# rowan.api_key = ""
result = rowan.submit_irc_workflow(
name="HNCO + H₂O - IRC",
initial_molecule=Molecule.from_xyz("""\
N -0.15519741 -1.36979175 -0.20679433
C 1.11565384 -1.23943631 -0.14797646
O 2.17614993 -1.72950370 -0.04017850
H -0.55869366 -2.29559315 -0.23834737
O 1.02571386 0.42871733 -0.27925360
H -0.09029954 -0.04166676 -0.31495768
H 1.26740151 0.88347299 0.53620841
"""),
method="GFN2-xTB",
)
print(f"View privately on the web at: https://labs.rowansci.com/irc/{result.uuid}")
result = result.wait_for_result().fetch_latest()
print(result)
Further Reading
If you're interested in exploring Rowan's intrinsic reaction coordinate and other reactivity tools, make a free account and start running calculations in minutes!
