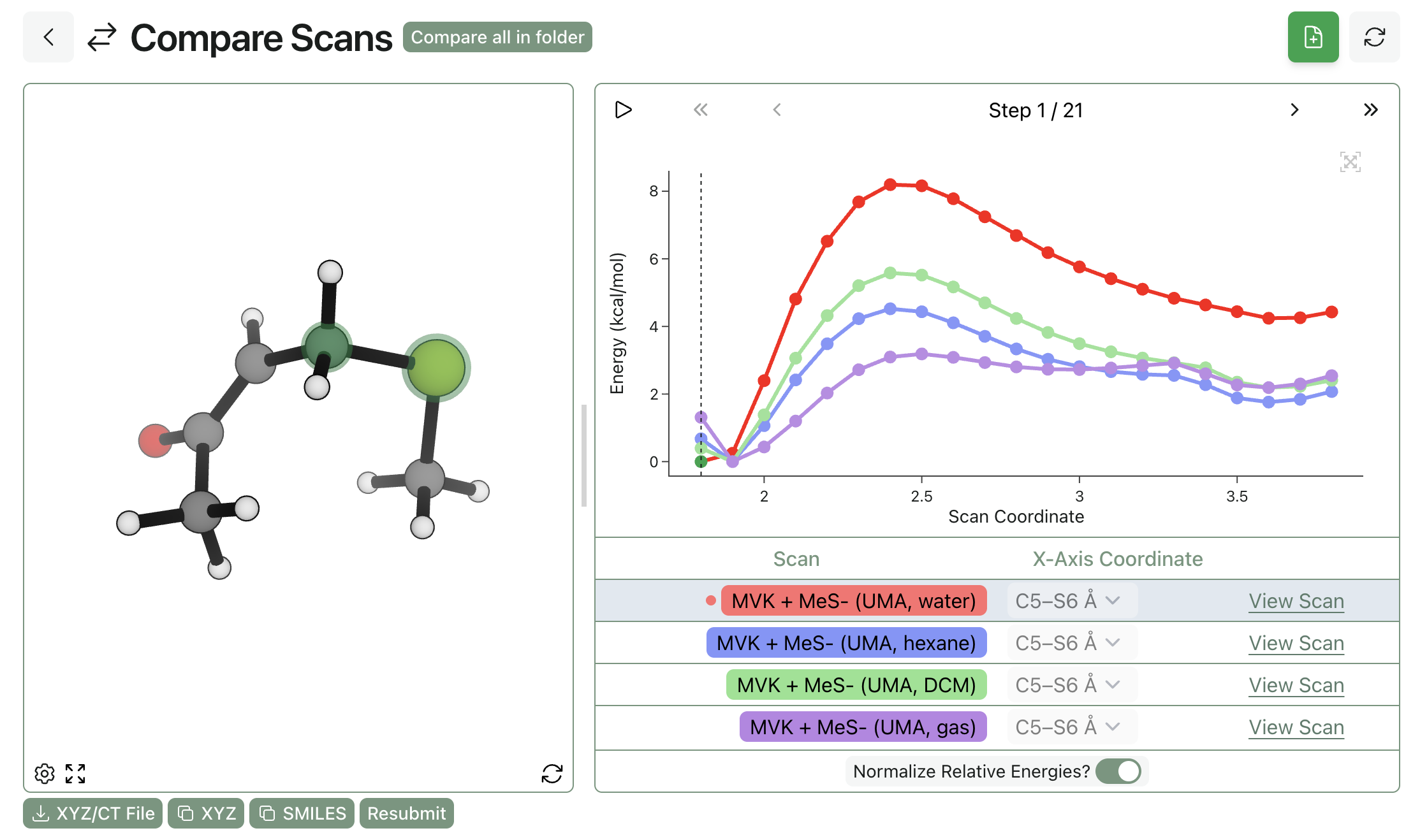
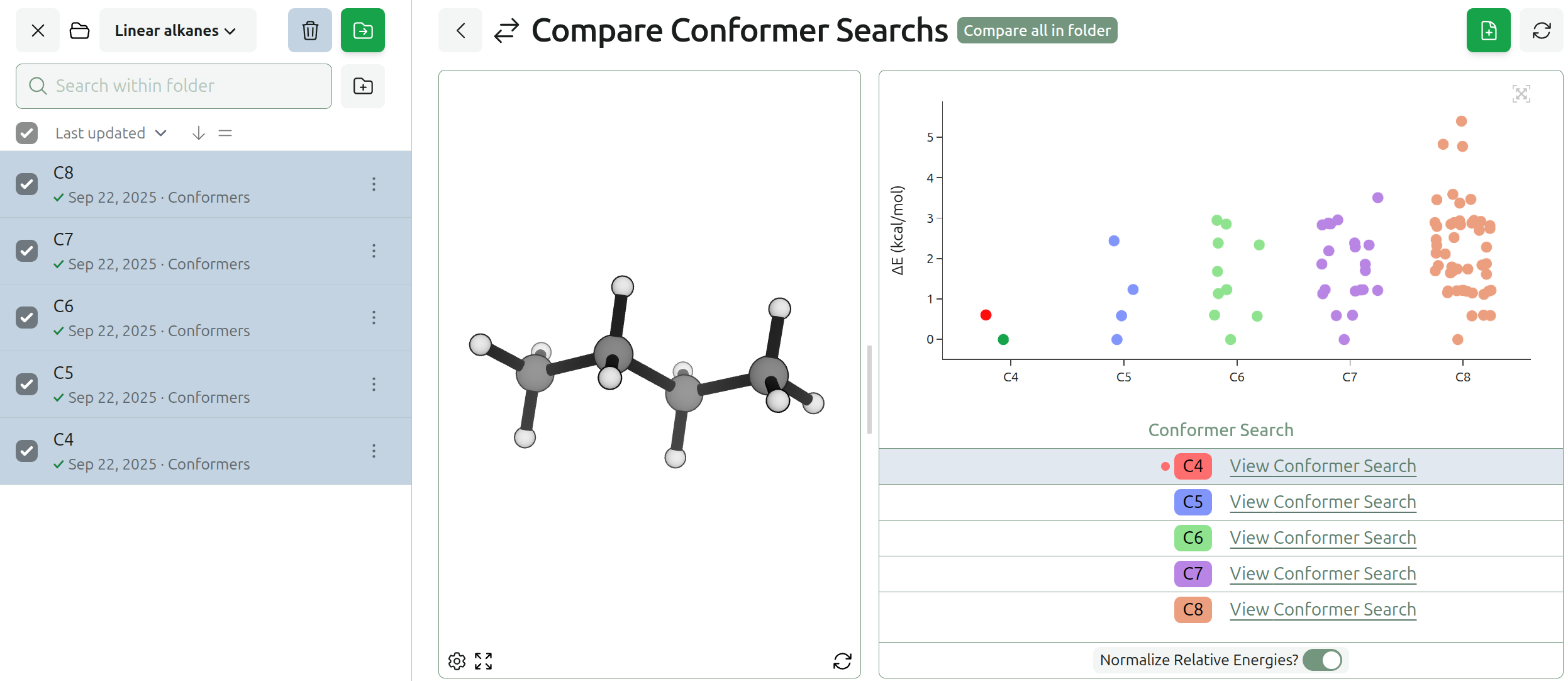
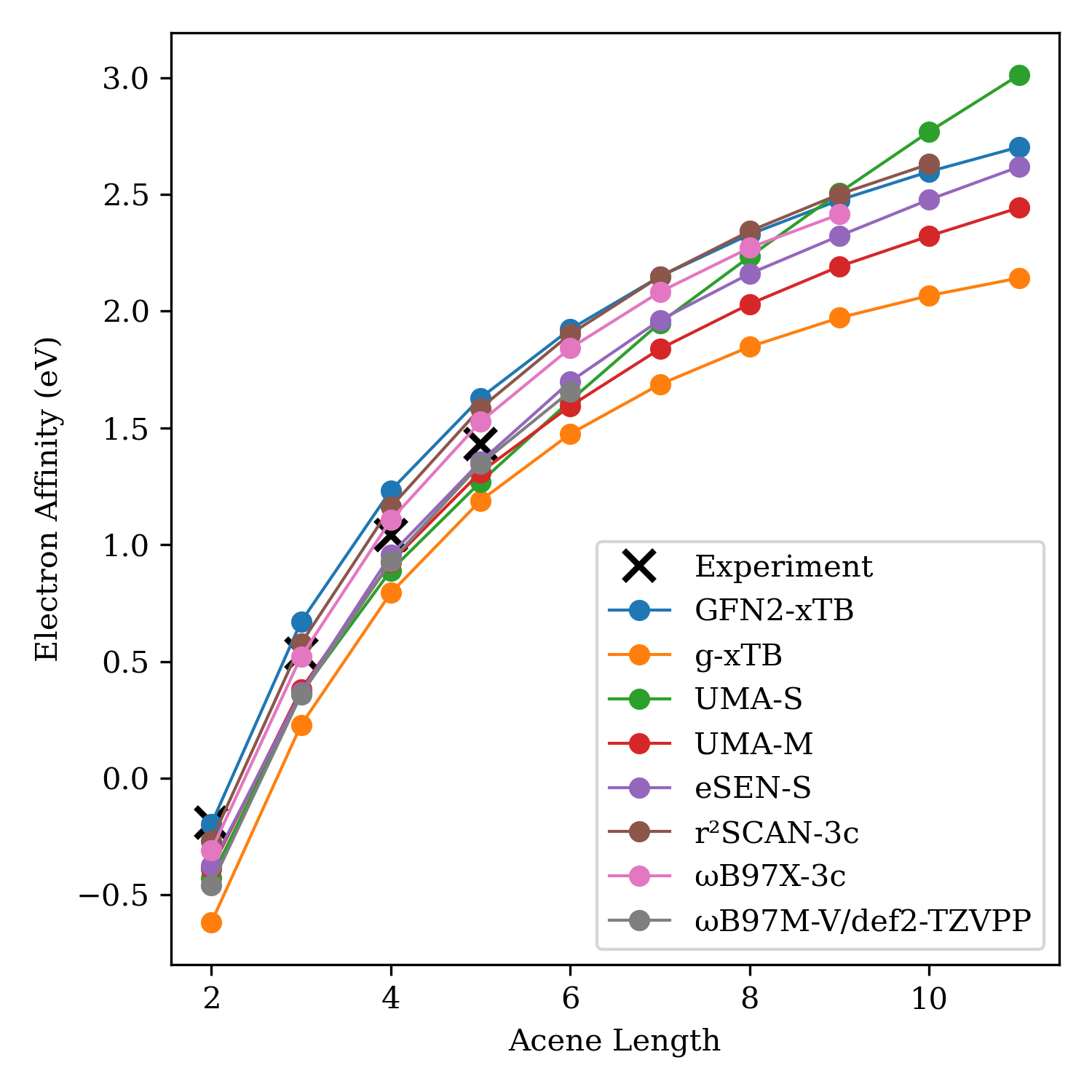
What Rowan Learned From the NVIDIA Atomistic Simulation Summit
by Corin Wagen and Spencer Schneider · Oct 9, 2025
This week, we had the pleasure of attending the NVIDIA Atomistic Simulation Summit, a two-day event held at NVIDIA's headquarters in Santa Clara. The summit brought together about a hundred researchers and engineers working at the frontier of atomistic simulation, from application developers like us to library builders and people working on low-level compute hardware. In keeping with the current state of the field, a lot of talks discussed neural network potentials (NNPs): how to train them, how to efficiently run inference, and how they could be used to great effect in practical simulations.

Contemplating existence inside NVIDIA's Endeavor building.
There were talks on a wide variety of interesting topics. To name just a few:
- Stephen Jones, one of the lead CUDA architects, spoke about upcoming developments to the CUDA ecosystem and how they might impact scientific software developers.
- Aditi Krishnapriyan shared her group's work on model distillation, transformer-based NNP architectures, and new approaches for transition-path sampling.
- Gianni de Fabritiis talked about integrating NNPs into free-energy perturbation workflows (both the good and the bad).
- Sebastian Ehlert explained Skala, a modern machine-learned density functional that's far more accurate than previous non-hybrid functionals.
- Marcel Müller described the use of large language models (LLMs) to drive quantum chemistry workflow through the El Agente framework.
- And Peter Eastman talked about his efforts to efficiently incorporate NNPs into OpenMM.
(There were many other excellent talks, but a full list would make this blog post a bit too long.)
Rowan's Presentation
Rowan was also invited to give a presentation to the NVIDIA audience. We talked about our work using NNPs and other modern computational techniques to drive real-world impact for problems in drug discovery and materials science. Our perspective is a bit different than most of the attendees, since we're able to directly work with a lot of customers on interesting scientific problems, so we tried to bring some "field notes" on how these models were being used right now (and where they weren't).

Corin's talk at NVIDIA.
We also did our best to state what we felt were controversial or underrated truths in the field. Here's a few:
- NNPs might eventually become a replacement for forcefields, but they can be a replacement for DFT right now. Too many papers focus on the former, in our opinion—there's a lot to do that doesn't require <10 ms inference times.
- Zero-shot inference is almost always better than fine-tuning because it can be benchmarked. We're able to deploy models like AIMNet2, Egret-1, and UMA to customers with confidence because we've run hundreds of thousands of calculations—fine-tuning always has the possibility to introduce additional subtle errors, and the requisite benchmarking is hard and time-consuming.
- Contra what many in the field are saying, we think that there's a value to doing the exact same science with NNPs that could previously be done with density-functional theory. Quantity has a quality of its own, and making calculations 10 or 100 times faster opens the door to entirely new kinds of research, like automatically computing strain for every docking pose or quickly standardizing libraries of SMILES strings.
- Perhaps most controversial of all: in the future, we think that all chemists will be computational chemists (at least a little). Library and application developers need to prepare for a future where the majority of their users may be experimentalists (or AI agents, although that's a different discussion)—and this should be seen as a victory for our field, not a threat.
What We Learned
While we don't feel we can share any of the exciting unpublished results we saw (sorry), here's some of our higher-level takeaways from the conference.
-
Bridging the gap between nanoscale and mesoscale is incredibly important. A number of talks discussed how to use atom-scale insight and featurization to model mesoscale phenomena, like solid–liquid interfaces, complex polymers in solution, or gas–solid reactions. These systems often have over 100,000 atoms and are complicated to prepare, simulate, and analyze. Still, the complexity of such systems is often unavoidable—the complex interactions between various mixture components and phases are part of what make these systems interesting, and these interactions cannot easily be reproduced in simpler model systems.
-
Interoperability matters. Both for AI agents and for human scientists, being able to quickly and smoothly combine results from many different underlying packages and scientific paradigms is key to building practically useful models. The best science rarely comes from a single model. Instead, skilled practitioners understand the strengths and limits of different approaches and combine them to get state-of-the-art results. Since very few scientific codes prioritize interoperability, getting the engineering right can be very difficult.
-
GPUs are getting bigger. Modern GPUs are large and have a ton of memory and parallel ability. It is hard to use all the capacity of the GPU efficiently for many tasks—for instance, using NNPs with small organic molecules doesn't even come close to saturating an H100 or H200. This leads us to our next point…
-
Batching matters. GPUs are designed for parallel execution; when using a GPU, it's almost always more efficient to run calculations in parallel (where possible). Unfortunately, this creates complex software-design challenges. Batching can be done at many levels: kernel batching, library-level batching, application-level batching, and so on. An MD code might be forced to decide whether to batch at a high level (running multiple replicates at once) or a lower level (batching neighbor-list class or force evaluations), and these choices are decidedly non-trivial from an architecture perspective.
-
Integrating ML into C++ Applications is difficult. There's not a simple way to integrate a PyTorch model into an existing scientific package written in C++. TorchScript is historically one of the more popular methods—but TorchScript is now deprecated, and models in TorchScript cannot AFAIK take advantage of libraries like cuEquivariance. (The replacement for TorchScript,
torch.export(), is not yet stable enough for production usage.) Simple architectures can be rewritten wholesale in C++, but this quickly becomes burdensome. Calling Python from C++ can work, but this creates all sorts of communication headaches. To our knowledge, no satisfactory solution exists here. -
Many old scientific packages will have to adapt to the GPU age. Most of the venerable molecular dynamics and quantum chemistry packages were not written with modern hardware in mind. It can be very difficult to get legacy C++ or FORTRAN code to work well on new GPUs, and considerable effort is needed to migrate these code bases—in many cases, wholesale rewrites may prove easier. This is painful but likely necessary for the field to mature.
-
The optimal architecture for NNPs remains unclear. There have been many contentious debates in the literature about NNP architectures: some authors use explicit long-range physical forces and rotational equivariance, others prefer to implicitly learn physics through a highly expressive equivariant architecture and still other researchers allow models to learn equivariance implicitly from the data. Despite a plethora of studies, no clear conclusion about which is best yet exists. Recent work from Aditi Krishnapriyan suggests that a simple transformer can even suffice in place of a graph neural network (vide supra), a result which we frankly find shocking. (Some of our team has written about these issues before: see Corin's blog post on long-range forces in NNPs.)
We're happy to be a part of the NVIDIA ecosystem, and we're grateful for all the work that the NVIDIA team is doing to advance open science and help translate advances in computing hardware to the simulation ecosystem. Looking forward to future summits!